To date, the exact mechanism behind many life processes has remained elusive, as our methods of "seeing" the tiny parts of proteins and how they interact have been limited. Now, a new, hybrid technique harnesses the power of the electron microscope, nanotechnology, sophisticated image processing and 3D mapping software, and cutting-edge methods of marking and measuring proteins. The breakthrough method is revealing the architecture and function of our tiniest moving parts, beginning with the physical initiation of reading DNA itself.
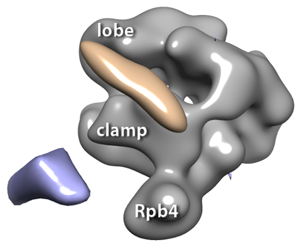
3D reconstructions of RNA polymerase II complexes (in gray) with 1.8 nm Ni-NTA-Nanogold®: blue volume indicates the Au when it is labeled at the C-terminus of Rpb4, the fourth subunit of RNA polymerase II, and the pink volume indicates the Au when it is labeled at the C-terminus of Tfg2, the second subunit of TFIIF.
Vital to the expression of a cell's genes is transcription, the "reading off" of DNA sections to begin creating needed proteins. A key player is RNA polymerase II, or RNAPII, a nano-machine that synthesizes messenger RNA from the starting DNA. Equally important is its sidekick, TFIIF, which can recognize DNA promoters and signal to RNAPII where to begin reading within a length of genetic code. So far, the mechanism behind the interaction of this dynamic duo has remained elusive, being too tiny to visualize.
In Taiwan, Dr. Wei-hau Chang and his team pioneered a hybrid technique that yielded an unprecedented view of the anatomy and function of these important protein complexes. Pushing beyond the boundaries of even cryo-EM and X-ray crystallography, they used a novel electron microscope imaging technique for single particle analysis, then took miniscule measurements with FRET analysis to further improve the precision of their mapping. What they found not only resolved the mystery of the placement of TFIIF on RNAPII, but also revealed that these are moving parts, dramatically shifting to engage DNA—giving us a first look at the physical initiation of gene transcription.
As they set out to study the architecture of the DNA-reading complex formed by TFIIF and RNAPII, Chang et al. needed to mark the hard-to-see proteins with a more visible tag, which could then be used to pin down their exact location. They began by tagging the end of the TFIIF protein with Nanogold®, a single gold nanoparticle; gold is electron-dense, and so shows up brightly in the electron microscope. The team used gene expression techniques in yeast to create a TAP tag on TFIIF with 10 histidines, which then bound tightly to Nickel-NTA-Nanogold® (Nanoprobes Inc.), giving them a clear marker for EM work.
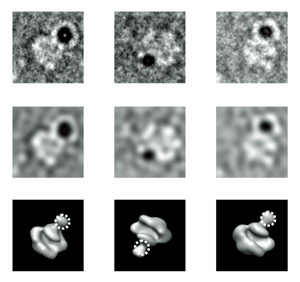
TEM projections of RNA polymerase II-TFIIF labeled with 5 nm Ni-NTA-Nanogold®: raw images in first row, filtered images in the second and the corresponding views of RNA polymerase II.
The first stage of analysis combined two imaging methods with the electron microscope. A more defocused view actually adds contrast to larger items, giving shape to the protein complex as a whole; for this set of images, the group used larger, 5 nm Ni-NTA-Nanogold markers. Next, a highly focused image was used to clearly pick out the single, tiny gold nanoparticle; here, the group used 1.8 nm Ni-NTA-Nanogold to mark their target with greatest precision. Together, the two views formed an image that shows the location of the tiny TFIIF transcription initiator, and also how it fits into the larger RNAPII transcription machine. Thousands of image pairs were analyzed by software to create a detailed 3D map of the proteins.
A second stage of analysis brought even greater precision, using FRET analysis. Here, a fluorescent "transmitter" marker was placed at a stable point on the RNAPII transcription machine, with an "acceptor" marker placed on the TFIIF protein. After a period of time, the amount of energy collected by the acceptor could be used to calculate its exact distance from the transmitter. The analysis of many samples further refined the localization of the TFIIF protein on the RNAPII complex, creating a detailed map in combination with the EM data.
Chang's team went much farther than simply mapping the architecture of these proteins in isolation, however. They also presented the pre-initiation transcription complex with gene sequences, and used their method to check the positioning of the TFIIF initiator as it encountered a DNA promoter. They found that the TFIIF makes a dramatic physical movement in response, which actually moves the DNA toward the lobe/protrusion domains of the RNAPII transcription machine. This likely represents the kinetic selection of a transcription starting position, and is a first view of the mechanism behind the start of a fundamental life process, observed in action.
"We believe such approach is generally useful for studying the architecture and dynamics of many protein complexes," write the authors, who likened their two-step process to "zooming in with google maps." Indeed, Chang's group has found a way to "see" the incredibly tiny parts of our vital nano-machinery, allowing real understanding of their interaction and function as they work to create and support life itself.
"This new technique combining several technologies really opens the door to a much deeper understanding of how cellular processes work," said Dr. James F. Hainfeld, chief scientist at Nanoprobes, makers of the Ni-NTA-Nanogold® used in Chang's technique. "This is a great tool for mapping protein structures at high resolution."

From right to left: Dr. Wei-hau Chang, Dr. Jen-wei Chang (the first author of Journal of Structural Biology 184, 52-62), Dr. Yi-min Wu (second author), Dr. Chun-hsiung Wang, Dr. Jheng-syong Wu, and Rob S.-H. Huang (Ph. D. candidate).
Read the scientific paper online:
Jen-Wei Chang, Yi-Min Wua, Zi-Yun Chen, Shih-Hsin Huang, Chun-Hsiung Wanga, Pei-lun Wua, Yi-ping Wenga, Changjiang You, Jacob Piehler, Wei-hau Chang. Hybrid electron microscopy-FRET imaging localizes the dynamical C-terminus of Tfg2 in RNA polymerase
II–TFIIF with nanometer precision. J. Struct. Biol. (2013).