![[Nanoprobes Tech Support (14k)]](../Images/logo_tech.gif)
Updated: January 15, 2010 (web)
5 nm Ni-NTA-Nanogold®
[5 nm Ni-NTA-Nanogold® product page]

| Product Name: |
5 nm Ni-NTA-Nanogold® |
| Catalog Number: |
2082 |
| Appearance: |
Clear red solution |
Revision: |
1.0 (December 2009) |
| Size: |
3 mL |
| Storage:: |
Upon receipt store product at 2 - 8°C. Product is shipped at ambient temperature. |
Technical Assistance Online
![[2080-PDF]](../Images/pdf.gif) Instructions (PDF) Instructions (PDF)
![[2080-PDF]](../Images/pdf.gif) Material Safety Data Sheet (PDF) Material Safety Data Sheet (PDF)
5 nm Ni-NTA-Nanogold® 1 is designed for the detection or localization of Histidine (His)-tagged recombinant proteins in multisubunit protein complexes, tissue or cell samples using transmission electron microscopy (TEM), scanning electron microscopy (SEM) and cryo-electron microscopy. Using 5 nm Ni-NTA-Nanogold®, His-tagged recombinant proteins originating from a variety of expression vectors can be labeled under both non-denaturing and denaturing conditions. The labeled His-tagged recombinant proteins can be directly visualized or localized by electron microscope (EM) without the use of gold or silver enhancement.
Warning: For research use only. Not recommended or intended for diagnosis of disease in humans or animals. Do not use internally or externally in humans or animals. Non radioactive and non carcinogenic.
The His-tag, consisting of five to ten consecutive histidine residues, has been used for purification of proteins by immobilized metal-ion affinity chromatography (IMAC).2,3 The use of a His tag provides several advantages. There is minimal addition of extra amino acids to the recombinant proteins. The small histidine tail is poorly immunogenic and usually does not interfere with protein folding. His-tagged proteins can have an extremely high affinity for metal ions (Ka=1013 M),3-6 which allows the detection of the His-tagged proteins using Ni-NTA (nickel (II) nitrilotriacetic acid) gold nanoparticles (catalog numbers 2080 and 2082).7-13 5 nm Ni-NTA-Nanogold® consists of a 5 nm gold particle with multiple Ni-NTA functionalities incorporated into ligands on the surface of gold particles. Each Ni2+ coordinates with one NTA and two histidines from the His-tagged recombinant protein to form a stable complex (Figure 1). A tight binding is achieved when three adjacent Ni-NTA groups bind to a 6x-His tag. The His-tagged proteins are thus labeled, and the 5 nm gold particles can be clearly visualized by electron microscopy without the use of gold or silver enhancement. Compared with immunogold labeling, the use of 5 nm Ni-NTA-Nanogold® or 1.8 nm Ni-NTA-Nanogold® provides the advantage of more precise localization of the His tag sites, since no additional protein entities or antibodies are involved and the distance from the gold particle to the His tag is less than 1.5 nm. The 5 nm Ni-NTA-Nanogold® and 1.8 nm Ni-NTA-Nanogold® bind to N-terminal, C-terminal and internal His-tag sequences, and recognize from five to ten consecutive histidine residues encoded by a variety of commercial available expression vectors.
Contents
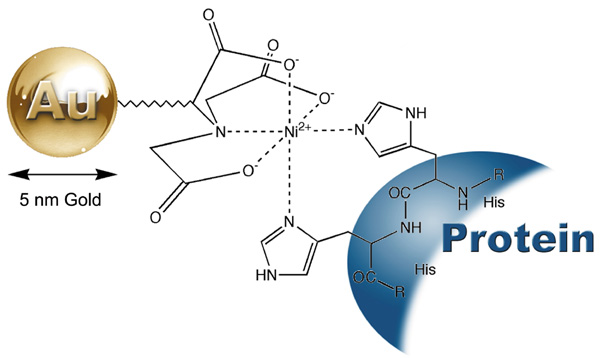
Figure 1: Interaction between a His-tagged protein and 5 nm Ni-NTA-Nanogold® (Catalog No.2082).
This product is supplied as a dark red colored liquid at a concentration of 0.5 µM in 50 mM MOPS, pH 7.9. No additional stabilizer or preservative is included. If a sterile solution is needed, filter the product with a 0.2 ?m cellulose acetate membrane filter. As supplied, this product is stable at least 1 year when stored at 2-8°C.
- His-tagged protein samples should be prepared in a binding buffer at pH 78. While His tags are labeled with 5 nm Ni-NTA-Nanogold®, nonspecific binding can also occur as many proteins have intrinsic histidine and/or cysteine amino acid residues. It is often necessary to optimize binding and washing conditions by varying the concentration of imidazole and sodium chloride in the binding and wash buffers. Increasing the concentration of imidazole and sodium chloride generally decreases nonspecific binding, but also weakens affinity interaction. The key is to find the right balance. 520 mM imidazole and 150 500 mM NaCl were generally found to give satisfactory labeling results. Including 1% (w/v) nonfat dry milk in the binding buffer and 0.1% (w/v) Tween®-20 in the binding/washing buffers especially helps reduce non-specific binding in tissue and cell samples. Buffers with low pH (<5.0) protonate the histidines and disrupt the interaction with the metal, and therefore should not be used.
- The binding buffer must be free from thiols such as beta-mercaptoethanol, or reducing or chelating agents such as DTT, EDTA or citrate. Samples containing EDTA, DTT, or citrate may give low specific staining.
- His tagged protein samples should be incubated with 5 nm Ni-NTA-Nanogold®, diluted 1/5 1/100 in the binding buffer for 5 -30 min at room temperature. The optimum concentration of 5 nm Ni-NTA-Nanogold® to be used needs to be determined for each application. His tagged protein complexes can be immobilized on a glow-discharged, carbon-coated electron microscopy grid prior to labeling. The grid can then be incubated upside-down on a droplet of label solution on parafilm. Protein complexes can also be labeled in solution. However, the gold-to-His ratio and concentrations must be carefully controlled to avoid the formation of aggregates, since each gold particle contains multiple Ni-NTA groups. Using a 5- to 10-fold molar excess of 5 nm Ni-NTA-Nanogold® in a diluted mixture can help reduce the formation of aggregates. After incubation, the excess 5 nm Ni-NTA-Nanogold® can be removed by gel filtration, or ion exchange liquid chromatography, dialysis or centrifugation.
- For applications involving tissues or cells, block the sample with 20 mM Tris, 150 mM NaCl, 0.1% Tween® 20 at pH7.6 containing 1-5% Nonfat dried milk for 5-30 min at room temperature prior to the application of 5 nm Ni-NTA-Nanogold®. This blocking step can block some non-specific protein binding, and minimizes non-specific interactions.
- Stain with osmium tetroxide, uranyl acetate, lead acetate, tungsten- or vanadium-based negative stains to give contrast between different structures. The tungsten- (Nano-WTM, Catalog number 2018) or vanadium- (Nano-Van™, Catalog number 2011) based negative stains are specially useful for staining small structures such as multisubunit protein complexes. NanoVanTM gives a lighter stain than uranium, lead or tungsten-based negative stains, and allows easier visualization of 2-5 nm gold nanoparticles.14
Contents
Note: The following protocols are general examples of applications for this product. Specific experiments may require optimization.
- TBS: 20 mM Tris at pH 7.6 with 150 mM NaCl.
- PBS: 20 mM sodium phosphate at pH 7.4 with 150 mM NaCl.
- PBS-BSA: 20 mM sodium phosphate at pH 7.4 with 150 mM NaCl containing 0.5% (w/v) BSA and 0.1% (w/v) gelatin (high purity). Nonfat dried milk can be used to replace BSA. Including 0.5 M NaCl and 0.05% (v/v) Tween20 helps reduce nonspecific binding.
- PBS-1% BSA: 20 mM sodium phosphate at pH 7.4 with 150 mM NaCl containing 1.0% (w/v) BSA and 0.1% (w/v) gelatin (high purity). It has been found that replacing BSA with nonfat dried milk helps reduce background or nonspecific binding.
- Prepare protein complex in 20 mM Tris at pH 7.6 with 150 mM NaCl.
- Incubate protein complex with 10 molar excess of 5 nm Ni-NTA-Nanogold® for 30 minutes at room temperature or 4°C.
- Remove the unbound gold nanoparticles from labeled protein conjugates using gel filtration, or ion exchange chromatography, centrifugation or dialysis. The gel filtration chromatography media such as GE Healthcare Superose and Superdex and Bio-rad Bio-Gel are selected based on the molecular weight of the proteins and the preferred fractionation range. Concentrate the reaction mixture to a suitable volume for injection using membrane centrifugation (e.g. Amicon Ultra-4, Millipore). Elute with TBS. The first colored peak or shoulder is the conjugate while the second dark colored band is excess gold namoparticles.
- If desired, remove salt using membrane centrifugation or dialysis.
- Load gold labeled protein complex on a carbon coated EM grid.
- Perform negative stain such as Nano-Van™ (catalog number 2011), according to product instructions before examination.
- Load purified His-tagged protein complex on glow-discharged carbon coated EM grids, and remove excess liquid using filter paper.
- Place grid upside-down on a droplet of 5 nm Ni-NTA-Nanogold® and incubate for 30 minutes at room temperature.
- If desired, wash the grid upside-down on a droplet of 20 mM Tris at pH 7.6 with 150 mM NaCl containing 8 mM imidazole for 1 min at room temperature.
- Rinse with water.
- Perform negative stain such as Nano-Van™ (catalog number 2011), according to product instructions before examination.
Contents
Labeling tissues or cells before embedding and sectioning (the pre-embedding method)15,16 is used for the study of surface targets. It gives good preservation of cellular structure, and subsequent staining usually produces high contrast for study of the cellular details.
Cells in Suspension
- Optional fixing of cells: e.g., with glutaraldehyde (0.05 - 1% for 15 minutes) in PBS. Do not use Tris buffer since this contains an amine. After fixation, centrifuge cells (e.g. 1 mL at 107 cells/ml) at 300 X g, 5 minutes; discard supernatant; resuspend in 1 mL PBS. Repeat this washing (centrifugation and resuspension) 2 times.
- Incubate cells with 0.02 M glycine in PBS (5 mins) to quench the remaining aldehyde. Centrifuge, then resuspend cells in PBS-BSA buffer (specified above) for 5 minutes.
- Wash cells using PBS-BSA using repeated centrifugation and resuspension as described in step 1 (2 X 5 mins). Resuspend in 1 mL PBS-BSA.
- Place 50-200 µL of cells into Eppendorf tube. Dilute 5 nm Ni-NTA-Nanogold® in PBS-BSA buffer and add 30-50 L to cells; incubate for 15-30 minutes with occasional shaking.
- Wash cells in PBS-BSA by repeated centrifugation and resuspension as described in step 1 (2 X 5 mins). 5-20 mM imidazole may be included if nonspecific binding is concerned.
- Fix cells using a final concentration of 1% (v/v) glutaraldehyde in PBS for 15 minutes. Then remove fixative by washing with PBS (3 X 5 mins).
- Rinse in deionized water (2 X 5 min).
- Dehydrate and embed according to usual procedure. Use of a low-temperature resin (e.g. Lowicryl) is recommended.
- Stain with uranyl acetate, or lead citrate or other positive staining reagent before examination.
Tissue Sections
- Float on a drop of water for 5-10 minutes.
- Incubate with PBS-1% BSA for 5 minutes to block non-specific protein binding
- Rinse with PBS-BSA (1 min).
- Incubate with 5 nm Ni-NTA-Nanogold® diluted 1/5 1/100 in PBS-BSA for 15-30 minutes at room temperature.
- Rinse with PBS containing 5 mM imidazole (1 min), then PBS (3 X 1 min).
- Postfix with 1% glutaraldehyde in PBS (10 mins).
- Rinse in deionized water (2 X 5 min).
- Dehydrate and embed according to usual procedure. Use of a low-temperature resin (e.g. Lowicryl) is recommended.
- Stain with uranyl acetate, or lead citrate or other positive staining reagent before examination.
Contents
Labeling after embedding and sectioning (the post-embedding method)15,16 allows the access of Ni-NTA-Nanogold® to the interior of the cells or tissues, and is used to label both exterior and interior targets.
Note: Thin sections mounted on grids are floated on drops of solutions on parafilm or in well plates. Hydrophobic resins usually require pre-etching.
- Prepare sections on plastic or carbon-coated nickel grid. Float on a drop of water for 5 - 10 minutes.
- Incubate with PBS-1% BSA for 5 minutes to block non-specific protein binding sites.
- Rinse with PBS-BSA (1 min).
- Incubate with 5 nm Ni-NTA-Nanogold® diluted 1/5 1/100 in PBS-BSA for 15-30 minutes at room temperature.
- Rinse with PBS containing 5 mM imidazole (1 min), then PBS (3 X 1 min).
- Postfix with 1% glutaraldehyde in PBS at room temperature (3 mins).
- Rinse in deionized water for (2 X 5 min).
- Stain with uranyl acetate, or lead citrate or other positive staining reagent before examination.
Contents
- Reddy, V.; Lymar, E.; Hu, M., and Hainfeld, J. F.: 5 nm Gold-Ni-NTA binds His Tags. Microsc. Microanal., 11, (Suppl. 2: Proceedings) (Proceedings of Microscopy and Microanalysis 2005); Price, R.; Kotula, P.; Marko, M.; Scott, J. H.; Vander Voort, G. F.; Nanilova, E.; Mah Lee Ng, M.; Smith, K.; Griffin, P.; Smith, P., and McKernan, S., Eds.; Cambridge University Press, New York, NY, 2005, p. 1216CD (Paper).
- Porath, J., and Olin, B.: Immobilized metal ion affinity adsorption and immobilized metal ion affinity chromatography of biomaterials. Serum protein affinities for gel-immobilized iron and nickel ions. Biochemistry, 22, 1621-1630 (1983).
- Hochuli, E.; Bannwarth, W.; Döbeli, H.; Gentz, R., and Stber, D.: Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. Bio/Technology, 6, 1321-1325 (1988).
- Uhlén, M., and Moks, T.: Gene fusions for purpose of expression: an introduction. Methods Enzymol., 185, 129-143 (1990).
- Casey, J. L.; Keep, P. A.; Chester, K. A.; Robson, L.; Hawkins, R. E., and Begent, R. H. J.: Purification of bacterially expressed single chain Fv antibodies for clinical applications using metal chelate chromatography. J. Immunol. Meth., 179, 105-116 (1995).
- Schmitt, J.; Hess, H., and Stunnenberg, H. G.: Affinity purification of histidine-tagged proteins. Molecular Biology Reports, 18, 223-230 (1993).
- Wolfe, C. L.; Warrington, J. A.; Treadwell, L., and Norcum, M. T.: A three-dimensional working model of the multienzyme complex of aminoacyl-tRNA synthetases based on electron microscopic placements of tRNA and proteins. J. Biol. Chem., 280, 38870-38878 (2005).
- Bumba, L.; Tichy, M.; Dobakova, M.; Komenda, J., and Vacha, F.: Localization of the PsbH subunit in photosystem II from the Synechocystis 6803 using the His-tagged NiNTA Nanogold labeling. J. Struct. Biol., 152, 28-35 (2005).
- Buchel, C.; Morris, E.; Orlova, E., and Barber, J.: Localisation of the PsbH subunit in photosystem II: a new approach using labelling of His-tags with a Ni(2+)-NTA gold cluster and single particle analysis. J. Mol. Biol., 312, 371-379 (2001).
- Hainfeld, J. F.; Liu, W.; Halsey, C. M. R.; Freimuth, P., and Powell, R. D.: Ni-NTA-Gold Clusters Target His-Tagged Proteins. J. Struct. Biol., 127, 185-198 (1999).
- Promnares, K.; Komenda, J.; Bumba, L.; Nebesarova, J.; Vacha, F., and Tichy, M.: Cyanobacterial Small Chlorophyll-binding Protein ScpD (HliB) Is Located on the Periphery of Photosystem II in the Vicinity of PsbH and CP47 Subunits. J. Biol. Chem., 281, 32705-32713 (2006).
- Jiang, Z. G.; Simon, M. N.; Wall, J. S., and McKnight, C. J.: Structural analysis of reconstituted lipoproteins containing the N-terminal domain of apolipoprotein B. Biophys. J., 92, 4097-4108 (2007).
- Adami, A.; Garcia-Alvarez, B.; Arias-Palomo, E.; Barford, D., and Llorca, O.: Structure of TOR and its complex with KOG1. Mol. Cell., 27, 509-516 (2007).
- Tracz, E., Dickson, D. W., Hainfeld, J. F., and Ksiezak-Reding, H.: Paired helical filaments in corticobasal degeneration: the fine fibrillary structure with NanoVan. Brain Res., 773, 33-44 (1997); Gregori, L., Hainfeld, J. F., Simon, M. N., and Goldgaber, D.: Binding of amyloid beta protein to the 20S proteasome. J. Biol. Chem., 272, 58-62 (1997); Hainfeld, J. F.; Safer, D.; Wall, J. S.; Simon, M. N.; Lin, B. J., and Powell, R. D.; Methylamine Vanadate (NanoVan) Negative Stain. Proc. 52nd Ann. Mtg., Micros. Soc. Amer.; G. W. Bailey and Garratt-Reed, A. J., (Eds.); San Francisco Press, San Francisco, CA, 1994, p. 132.
- J. E. Beesley, in Colloidal Gold: Principles, Methods and Applications, M. A. Hayat, ed., Academic Press, New York, 1989; Vol. 1, pp. 421-425.
- Lujan, R.; Nusser, Z.; Roberts, J. D. B.; Shigemoto, R.; Ohishi, H., and Somogyi, P.: Differential plasma membrane distribution of metabotropic glutamate receptors mGluR1 alpha, mGluR2 and mGluR5, relative to neurotransmitter release sites. J. Chem. Neuroanat., 13, 219-241 (1997).
Technical Assistance Available.
For a complete list of references citing this product, please visit our References page.
Contents
|