![[Nanoprobes Tech Support (14k)]](../Images/logo_tech.gif)
Updated: January 15, 2010 (web)
1.8 nm Ni-NTA-Nanogold®
[1.8 nm Ni-NTA-Nanogold® product page]

| Product Name: |
1.8 nm Ni-NTA-Nanogold® |
| Catalog Number: |
2080 |
| Appearance: |
Brown solution |
Revision: |
2.1 (December 2009) |
| Size: |
3 mL |
| Storage:: |
Upon receipt store product at 2 - 8°C. Product is shipped at ambient temperature. |
Technical Assistance Online
![[2080-PDF]](../Images/pdf.gif) Instructions (PDF) Instructions (PDF)
![[2080-PDF]](../Images/pdf.gif) Material Safety Data Sheet (PDF) Material Safety Data Sheet (PDF)
1.8 nm Ni-NTA-Nanogold® is designed for detection or localization of histidine (his)-tagged fusion proteins using electron microscopy, light microscopy, blotting or other detection method. Using 1.8 nm Ni-NTA-Nanogold®, his-tagged fusion proteins originating from any of a variety of expression vectors can be labeled under both non-denaturing and denaturing conditions. The labeled his-tagged fusion proteins can be visualized by microscope or eyes when used with or without gold or silver enhancement reagents, such as GoldEnhance EM (Catalog number 2113), GoldEnhance LM (Catalog number 2112), HQ Silver (Catalog Number 2012), or LI Silver (Catalog number 2013).
Warning: For research use only. Not recommended or intended for diagnosis of disease in humans or animals. Do not use internally or externally in humans or animals. Non radioactive and non carcinogenic.
The His-tag, consisting of five to ten consecutive histidine residues, has been used for purification of proteins by immobilized metal-ion affinity chromatography (IMAC).1-3 The use of a His tag has several advantages. There is minimal addition of extra amino acids to the recombinant proteins. The small histidine tail is poorly immunogenic and usually does not interfere with protein folding. His-tagged proteins can have an extremely high affinity for metal ions (Ka=1013 M).1,4-6 This affinity for metal ions allows for the detection of the fusion proteins using Ni-NTA (nickel-nitrilotriacetic acid) Nanogold® (Catalog Number 2080). 1.8 nm Ni-NTA-Nanogold® consists of a 1.8 nm Nanogold particle with multiple nickel-nitrilotriacetic acid functionalities incorporated into the ligands on the surface of gold particles. Each Ni2+ coordinates with one nitrilotriacetic acid and two histidines from the fusion proteins, and forms a stable complex (Figure 1). The his-tagged fusion proteins are therefore labeled, and detected by electron microscopy, light microscopy or blotting, when used with or without gold or silver enhancement reagents.
Contents
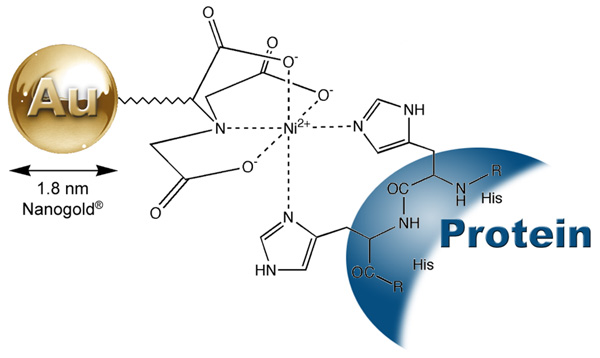
Figure 1: Interaction between a His-tagged protein and Ni-NTA-Nanogold®.
This product is supplied as a brown colored liquid at a concentration of 10 nmol/mL of Ni-NTA-Nanogold® in 50 mM MOPs, pH 7.9. No additional stabilizer or preservative is included. If a sterile solution is needed, filter the product with a 0.2 um cellulose acetate membrane filter. As supplied, this product is stable at least 1 year when stored at 2-8°C.
- For best results, prepare protein sample in a binding buffer of 20 mM Tris, 0.15 M NaCl, pH 7.6 with 0.1% (w/v) Tween® -20 and 1% (w/v) nonfat dried milk. Including Tween® -20 and nonfat dried milk helps reduce the non-specific binding of Ni-NTA-Nanogold® to the proteins. If significant amounts of non-specific staining are observed at the end of the detection, the concentration of sodium chloride in the binding buffer can be increased to 0.5 to 1 M.
- The binding buffer must be free from thiols such as beta-mercaptoethanol, or reducing or chelating agents such as DTT, EDTA or citrate. Samples containing EDTA, DTT, or citrate may give low specific staining.
- Add required amount of Ni-NTA-Nanogold®, diluted 1/5 1/100 in 20 mM Tris, 150 mM NaCl, 1% nonfat dried milk and 0.1% Tween 20 at pH 7.6, to the His-tagged proteins in the binding buffer. Incubate for 5 -30 minutes at room temperature. The optimum quantity of Ni-NTA-Nanogold® to be used needs to be determined for each application. Using five to ten fold molar excess of Ni-NTA-Nanogold® can help reduce the formation of cross-linking products. For most applications, 5-10 minute incubation is sufficient for obtaining a positive staining.
- After incubation, the excess Ni-NTA-Nanogold® can be removed by size exclusion liquid chromatography, dialysis or simply washed off from cell or tissue samples or membranes using a buffer containing 50 mM Tris, 300 mM NaCl, 10-200 mM imidazole and 0.1% Tween 20 adjusted to pH 7.6. The wash buffer may be optimized for specific applications by adjusting the imidazole and NaCl (up to 1M) concentration. Increasing the imidazole concentration helps reduce the non-specific interaction or background, and can also decease the binding to the target proteins. To determine the optimal combination of specific staining and background, different wash conditions should be tested.
- Buffers other than the suggested ones can be used if similar pH and ionic strength are used. In low pH (<5.0) buffers, histidines will be protonated, which disrupts their interaction with the metal.
- The Ni-NTA procedure can also be used under denaturing conditions. This can be useful in cases where the His-tag is inaccessible from the surface of the fusion proteins or if the protein is insoluble.
- For the application involving tissues or cells, block the sample with 20 mM Tris, 150 mM NaCl, 0.1% Tween 20 at pH 7.6 containing 1-5% Nonfat dried milk for 5-30 minutes at room temperature prior to the application of Ni-NTA-Nanogold®. The blocking step can block some non-specific protein binding sites and minimizes non-specific interaction.
- For most work, gold or silver enhancement with a neutral pH is recommended to give an enhanced signal for detection. The enhancement can be terminated by immersion of the sample or grid in deionized water or 2.5% of sodium thiosulfate for 1 minute as soon as satisfactory specific staining is reached. Further enhancement may lead to undesirable background. Applying gold or silver enhancement to a control sample without exposing to Ni-NTA-Nanogold® helps to determine the stability time when self-nucleation of the enhancement starts. Self-nucleation can generate high background. The actual enhancement time should never exceed the stability time.
- Because the 1.8 nm Nanogold® particles are small, over-staining with OsO4, uranyl acetate or lead citrate may tend to obscure direct visualization of individual Nanogold® particles. Therefore, the use of reduced amount or concentration of usual stains is recommended.
Contents
For most work, gold or silver enhancement is recommended to give a good signal in the electron microscope (see below). For particular applications, visualization of the Nanogold® directly may be desirable. Generally this requires very thin samples and precludes the use of other stains.
Nanogold® provides a much improved resolution and smaller probe size over other colloidal gold antibody products. However, because Nanogold® is only 1.8 nm in diameter, it will not only be smaller, but will appear less intense than, for example, a 5 nm gold particle. With careful work, however, Nanogold® may be seen directly through the binoculars of a standard EM even in 80 nm thin sections. However, achieving the high resolution necessary for this work may require new demands on your equipment and technique. Several suggestions follow:
- Before you start a project with Nanogold® it is helpful to see it so you know what to look for. Dilute the Nanogold® stock 1:5 and apply 4 µL to a grid for 1 minute. Wick the drop and wash with deionized water 4 times.
- View Nanogold® at 100,000 X magnification with 10 X binoculars for a final magnification of 1,000,000 X. Turn the emission up full and adjust the condenser for maximum illumination.
- The alignment of the microscope should be in order to give 0.3 nm resolution. Although the scope should be well aligned, you may be able to skip this step if you do step 4.
- Objective stigmators must be optimally set at 100,000 X. Even if the rest of the microscope optics is not perfectly aligned, adjustment of the objective stigmators may compensate and give the required resolution. You may want to follow your local protocol for this alignment but since it is important, a brief protocol is given here:
- At 100,000 X (1 X 106 with binoculars), over focus, under focus, then set the objective lens to in focus. This is where there is the least amount of detail seen.
- Adjust each objective stigmator to give the least amount of detail in the image.
- Repeat steps a and b until the in focus image contains virtually no contrast, no wormy details, and gives a flat featureless image.
- Now underfocus slightly, move to a fresh area, and you should see small black dots of 1.8 nm size. This is the 1.8 nm Ni-NTA-Nanogold®. For the 1:5 dilution suggested, there should be about 5 to 10 gold spots on the small viewing screen used with the binoculars. Contrast and visibility of the gold clusters is best at 0.2 - 0.5 m defocus, and is much worse at typical defocus values of 1.5 - 2.0 m commonly used for protein molecular imaging.
- In order to operate at high magnification with high beam current, thin carbon film over fenestrated holey film is recommended. Alternatively, thin carbon or 0.2% Formvar over a 1000 mesh grid is acceptable. Many plastic supports are unstable under these conditions of high magnification/high beam current and carbon is therefore preferred. Contrast is best using thinner films and thinner sections.
- Once you have seen 1.8 nm Ni-NTA-Nanogold® you may now be able to reduce the beam current and obtain better images on film. For direct viewing with the binoculars reduction in magnification from 1,000,000 X to 50,000 X makes the Nanogold® much more difficult to observe and not all of the golds are discernable. At 30,000 X (300,000 X with 10 X binoculars) Nanogold® particles are not visible. It is recommended to view at 1,000,000 X, with maximum beam current, align the objective stigmators, and then move to a fresh area, reduce the beam, and record on film.
- If the demands of high resolution are too taxing or your sample has an interfering stain, a very good result may be obtained using gold or silver enhancement to give particles easily seen at lower magnification.
Contents
1.8 nm Ni-NTA-Nanogold® will nucleate gold or silver deposition resulting in a dense particle 2-80 nm in size or larger depending on development time. The gold or silver enhancement is usually completed before applying any negative staining reagents such as osmium tetroxide, lead citrate or uranyl acetate, since these will nucleate gold or silver deposition in the same manner as gold and produce non-specific staining. Gold or silver development is recommended for applications of 1.8 nm Ni-NTA-Nanogold® in which these stains are to be used, otherwise the 1.8 nm Ni-NTA-Nanogold® particles may be difficult to visualize against the stain.
Protein samples or pecimens must be thoroughly rinsed with deionized water before silver enhancement reagents are applied. This is because the buffers used may contain chloride ions and other anions which form insoluble precipitates with silver. These are often light-sensitive and will give non-specific staining.
Fixing with osmium tetroxide may cause some loss of silver; if this is found to be a problem, slightly longer development time may be appropriate. Alternatively, use of 0.1% osmium tetroxide instead of 1% has been found to give similar levels of staining while greatly reducing etching of the silver particles.
Contents
His-tagged proteins can be blotted onto PVDF or nitrocellulose membranes directly or through an electrophoretic transfer, and detected by direct visualization using Ni-NTA-Nanogold® with silver or gold enhancement. The basic procedure is as follows:
- Spot or transfer his-tagged proteins onto PVDF or nitrocellulose membrane.
- Equilibrate membranes with 20 mM Tris, 150 mM NaCl, pH 7.6 and 0.1% (w/v) Tween® 20 (TBST) for 2 min.
- Block with 20 mM Tris, 150 mM NaCl, pH 7.6 containing 5% nonfat dried milk and 0.1 % Tween® 20 for 30 minutes at room temperature
- Rinse with TBST (3 X 5 mins).
- Incubate with 1.8 nm Ni-NTA-Nanogold® diluted 1 : 50 in 20 mM Tris, 150 mM NaCl, pH 7.6 containing 1% nonfat dried milk, and 0.1% Tween® 20 for 5 minutes at room temperature.
- Rinse with TBST and 10 mM imidazole (3 X 5 min). The concentration of imidazole and sodium chloride can be increased if high non-specific binding is observed.
- Rinse with deionized water (4 X 3 mins).
- Perform gold or silver enhancement as directed in the instructions for gold or silver reagents.
- Stop the enhancement when satisfactory staining is achieved. The enhancement can be stopped by washing membrane with deionized water (3 X 5 mins).
Contents
- Hochuli, E.; Bannwarth, W.; Döbeli, H.; Gentz, R., and Stber, D.: Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. Bio/Technology, 6, 1321-1325 (1988).
- Porath, J., and Olin, B.: Immobilized metal ion affinity adsorption and immobilized metal ion affinity chromatography of biomaterials. Serum protein affinities for gel-immobilized iron and nickel ions. Biochemistry, 22, 1621-1630 (1983).
- Hochuli, E.; Döbeli, H., and Schacher, A.: New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J. Chromatograph., 411, 177-184 (1987).
- Uhlén, M., and Moks, T.: Gene fusions for purpose of expression: an introduction. Methods Enzymol., 185, 129-143 (1990).
- Casey, J. L.; Keep, P. A.; Chester, K. A.; Robson, L.; Hawkins, R. E., and Begent, R. H. J.: Purification of bacterially expressed single chain Fv antibodies for clinical applications using metal chelate chromatography. J. Immunol. Meth., 179, 105-116 (1995).
- Schmitt, J.; Hess, H., and Stunnenberg, H. G.: Affinity purification of histidine-tagged proteins. Molecular Biology Reports, 18, 223-230 (1993).
- Kollman, J. M.; Zelter, A.; Muller, E. G.; Fox, B.; Rice, L. M.; Davis, T. N., and Agard, D. A.: The Structure of the gamma-Tubulin Small Complex: Implications of Its Architecture and Flexibility for Microtubule Nucleation. Mol. Biol. Cell, 19, 207-215 (2008).
- Kollman, J. M.; Zelter, A.; Muller, E. G.; Fox, B.; Rice, L. M.; Davis, T. N., and Agard, D. A.: The Structure of the gamma-Tubulin Small Complex: Implications of Its Architecture and Flexibility for Microtubule Nucleation. Mol. Biol. Cell, 19, 207-215 (2008).
- Balasingham, S. V.; Collins, R. F.; Assalkhou, R.; Homberset, H.; Frye, S. A.; Derrick, J. P., and Tonjum, T.: Interactions between the Lipoprotein PilP and the Secretin PilQ in Neisseria meningitidis. J. Bacteriol., 189, 5716-5727 (2007).
- Jiang, Z. G.; Simon, M. N.; Wall, J. S., and McKnight, C. J.: Structural analysis of reconstituted lipoproteins containing the N-terminal domain of apolipoprotein B. Biophys. J., 92, 4097-4108 (2007).
- Pye, V. E, Beuron, F, Keetch, C. A, McKeown, C, Robinson, C. V, Meyer, H. H, Zhang, X, and Freemont, P. S.: Structural insights into the p97-Ufd1-Npl4 complex. Proc. Natl. Acad. Sci. USA, 104, 467-472 (2007).
- Promnares, K.; Komenda, J.; Bumba, L.; Nebesarova, J.; Vacha, F., and Tichy, M.: Cyanobacterial Small Chlorophyll-binding Protein ScpD (HliB) Is Located on the Periphery of Photosystem II in the Vicinity of PsbH and CP47 Subunits. J. Biol. Chem., 281, 32705-32713 (2006).
- Collins, R. F.; Beis, K.; Clarke, B. R.; Ford, R. C.; Hulley, M.; Naismith, J. H.; and Whitfield, C.: Periplasmic protein-protein contacts in the inner membrane protein Wzc form a tetrameric complex required for the assembly of Escherichia coli group 1 capsules. J. Biol. Chem., 281, 2144-2150 (2006).
- Wolfe, C. L.; Warrington, J. A.; Treadwell, L., and Norcum, M. T.: A three-dimensional working model of the multienzyme complex of aminoacyl-tRNA synthetases based on electron microscopic placements of tRNA and proteins. J. Biol. Chem., 280, 38870-38878 (2005).
- Bumba, L.; Tichy, M.; Dobakova, M.; Komenda, J., and Vacha, F.: Localization of the PsbH subunit in photosystem II from the Synechocystis 6803 using the His-tagged NiNTA Nanogold labeling. J. Struct. Biol., 152, 28-35 (2005).
- Collins, R. F.; Frye, S. A.; Balasingham, S.; Ford, R. C.; Tonjum, T., and Derrick, J. P.: Interaction with type IV pili induces structural changes in the bacterial outer membrane secretin PilQ. J. Biol. Chem., 280, 18923-18930 (2005).
- Collins, R. F.; Frye, S. A.; Balasingham, S.; Ford, R. C.; Tonjum, T., and Derrick, J. P.: Interaction with type IV pili induces structural changes in the bacterial outer membrane secretin PilQ. J. Biol. Chem., 280, 18923-18930 (2005).
- Chatterji, A.; Ochoa, W. F.; Ueno, T.; Lin T., and Johnson, J. E.: A virus-based nanoblock with tunable electrostatic properties. Nano Lett., 5, 597-602 (2005).
- Hainfeld, J. F.; Liu, W.; Joshi, V., and Powell, R. D.: Nickel-NTA-Nanogold Binds his-Tagged Proteins. Microsc. Microanal., 8, (Suppl. 2: Proceedings); Voekl, E.; Piston, D.; Gauvin, R.; Lockley, A. J.; Bailey, G. W., and McKernan, S., Eds.; Cambridge University Press, New York, NY, 832CD (2002) (Paper).
- Buchel, C.; Morris, E.; Orlova, E., and Barber, J.: Localisation of the PsbH subunit in photosystem II: a new approach using labelling of His-tags with a Ni(2+)-NTA gold cluster and single particle analysis. J. Mol. Biol., 312, 371-379 (2001).
- Hainfeld, J. F.; Liu, W.; Halsey, C. M. R.; Freimuth, P., and Powell, R. D.: Ni-NTA-Gold Clusters Target His-Tagged Proteins. J. Struct. Biol., 127, 185-198 (1999).
Technical Assistance Available.
For a complete list of references citing this product, please visit our References page.
Contents
|