 |
Nanoprobes, Incorporated
95 Horseblock Road, Unit 1, Yaphank, NY 11980-9710
Tel: (877) 447-6266 (Toll-Free in US) or (631) 205-9490 | Fax: (631) 205-9493
tech@nanoprobes.com | www.nanoprobes.com |
Updated: November 15, 2006
NANOGOLD® PRODUCT INFORMATION
Monomaleimido Nanogold® Labeling Reagent
[Monomaleimido Nanogold® Labeling Reagent product page]

| Product Name: |
MONOMALEIMIDO NANOGOLD® |
| Quantity: |
6 nmol |
| Catalog Number: |
2020S |
| Appearance: |
Brown Powder/Solid |
| Revision: |
1.8 (November 2006) |
Technical Assistance Online
![[2020S-PDF]](../Images/pdf.gif) Instructions (PDF) Instructions (PDF)
![[2020S-PDF]](../Images/pdf.gif) Material Safety Data Sheet (PDF) Material Safety Data Sheet (PDF)
Congratulations on your acquisition of a revolutionary new gold immunoreagent: the MONOMALEIMIDO NANOGOLD® labeling kit. With this reagent you can label your own primary antibodies (IgG or Fab' fragments), or any other biomolecule containing an accessible sulfhydryl group, with NANOGOLD®. Because NANOGOLD® is a discrete molecular compound and not a colloidal gold preparation, conjugates prepared with this reagent have several advantages over colloidal gold conjugates (see below).
Guide to Gold Cluster Labeling: our detailed description of gold cluster labeling, how to isolate conjugates and calculate the extent of labeling, is now available, which includes spectra, micrographs, and tips and suggestions for better labeling (opens in a new window).
Warning: For research use only. Not recommended or intended for diagnosis of disease in humans or animals. Do not use internally or externally in humans or animals. Non radioactive and non carcinogenic.
NANOGOLD® is a newly developed gold label, prepared using a discrete gold compound rather than a colloid.1 This kit contains the NANOGOLD® particle with a single maleimide functionality incorporated into a ligand on the surface of the gold particle; this has a specific reactivity towards sulfhydryl groups, and may be covalently linked to reduced disulfides in the hinge region of antibodies, as shown in the figure below (Figure 1). It may also be used to label cysteine residues in proteins, or other biomolecules with an accessible sulfhydryl group. Molecules which have been labeled using Monomaleimido-NANOGOLD® include proteins,2 peptides,3 and oligonucleotides.4 The reagent as supplied has been lyophilized from 0.02 M sodium phosphate at pH 6.5, with 150 mM sodium chloride; dissolution in 1 ml deionized water will produce a solution of activated NANOGOLD® in this buffer. NANOGOLD® conjugates can be used in immunoblotting, light microscopy, and electron microscopy to provide clear visibility. They are stable to wide ranges of pH and ionic strength, and are not radioactive or carcinogenic. The labeling reagent should be stored at -20°C.
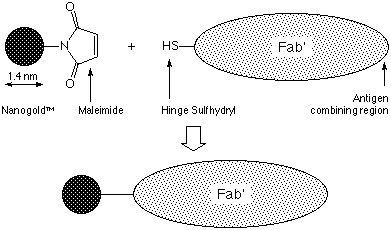 |
| Figure 1: Schematic showing NANOGOLD® labeling of Fab' fragment via reaction of a sulfhydryl and a maleimide group. |
Sufficient reagent is supplied to label up to 3 nmol of sulfhydryl groups; this corresponds to 0.15 mg of Fab' fragments, or 0.45 mg of IgG molecules.
Contents
NANOGOLD® particles degrade upon exposure to thiols such as beta-mercaptoethanol or dithiothreitol. Thiol compounds will also react with the maleimide group and render the particle non-reactive for conjugation. Thiol compounds used in the reduction of protein molecules (or other biomolecules) should be separated from the protein by gel filtration before NANOGOLD® conjugation. Use a gel, such as Matrex GH25 (Millipore), which has an exclusion cut-off at molecular weight 3,000. Dialysis does NOT provide acceptable purification in this application. A small amount of residual thiol reagent can severely limit the performance of NANOGOLD®.
Although NANOGOLD® is usually stable,5 under demanding conditions, including pH values lower than 4 or ionic strengths above 0.3 M, NANOGOLD® reagents, labeled specimens or conjugates may not be stable above 50°C, and best results are obtained at room temperature or 4°C. In such cases, avoid 37°C incubations, and use low temperature embedding media (e.g., Lowicryl) if labeling before embedding;6 do not bake tissue blocks with NANOGOLD®. If your experiment requires higher temperature embedding, then silver enhancement should be performed before embedding.
Contents
IgG molecules contain disulfide bonds which connect the chains in the hinge region. These are selectively reduced with a mild reducing agent, such as dithiothreitol (DTT) or mercaptoethylamine hydrochloride (MEA), then reacted with the NANOGOLD® reagent in buffer solution, either for 1 hour at room temperature or overnight at 4°C. The coupling reaction should be performed at pH 6.5, since at pH values greater than 7 the maleimido group becomes reactive towards primary amines as well as sulfhydryls, and may give non-specific labeling. Hydrolysis of the maleimide group also happens faster at higher pH. The reduced antibody must be isolated from the reducing agent before reaction; this may be achieved by gel filtration chromatography, using a gel such as Matrex GH25 (Millipore), which has an exclusion cutoff at molecular weight 3,000 . The NANOGOLD® conjugated product is separated from unlabeled NANOGOLD® by gel filtration chromatography, using a fine gel such as GE Healthcare (formerly Amersham Pharmacia) Superose 6 or 12, or Superdex 75. The recommended procedure is given below:
- Dissolve 0.3 mg of the IgG antibody (2 nmol) in 1 mL of 0.1 M sodium phosphate buffer, pH 6.0, containing 5 mM EDTA. If the IgG is already in solution, exchange into the phosphate/EDTA buffer using membrane centrifugation (e.g. Centricon-50 system, Millipore) or a desalting column. Add 1 mL of the IgG solution to a vial containing 6 mg of mercaptoethylamine hydrochloride (MEA) (Final concentration 0.05M). Incubate at 37°C for 90 minutes.
- Isolate reduced antibody by gel filtration chromatography. Use a gel, such as Matrex GH25 (Millipore), which has an exclusion cut-off at molecular weight 3,000. Dialysis does NOT provide acceptable purification in this application. As the eluent, use 0.02 M sodium phosphate at pH 6.5, with 150 mM sodium chloride and 1 mM EDTA. The reduced antibody will be eluted in the void volume as the first sharp peak in the trace. Combine the fractions containing reduced antibody; the total amount of antibody should be calculated from the optical density (usually for IgG, E1% at 280 nm = 14.5; concentration in mg/mL = OD280nm X 10/E1% = OD280nm X 0.69).
- Dissolve the NANOGOLD® reagent in 0.2 mL of deionized water. If the reagent is slow to dissolve, vortex the solution. Sufficient reagent is supplied to label up to 0.45 mg of IgG; if you are using a smaller amount, use a proportionately smaller amount of the NANOGOLD® reagent (e.g. half the reagent supplied will label up to 0.225 mg of IgG). Once the NANOGOLD® reagent is reconstituted with water, it should be used immediately. The maleimide group is hydrolyzed in aqueous solution.
- Add the reconstituted NANOGOLD® solution to the reduced antibody immediately. After mixing, adjust the reaction volume to give an IgG concentration of 2 nmol/mL (0.3 mg/mL). Incubate for 2 hours at room temperature. For an overnight reaction at 4°C, reduce the reaction volume to give an IgG concentration of 6 nmol/mL (9 mg/mL) using a membrane centrifugation device with a 10 kDa molecular weight cut-off (e.g. Centricon-10 system, Millipore).
- Separate the unbound gold particles from the antibody conjugates using gel filtration chromatography. The NANOGOLD® conjugate may be effectively isolated using a medium such as GE Healthcare (formerly Amersham Pharmacia) Superose 6 or 12 (which fractionate a wide range of molecular weights) or Superdex-75, or an alternative that effectively fractionates your protein from smaller species. Concentrate the reaction mixture to a suitably small volume using membrane centrifugation (e.g. Centricon-30 system, Millipore). Elute with 0.02 M sodium phosphate at pH 7.4 with 150 mM sodium chloride; the first, faintly colored peak is the conjugate, while the second, darker band is unbound NANOGOLD® particles. For even higher purity, repeat the process one time.
Estimation of NANOGOLD®/antibody ratio: Dilute a portion of the labeled antibody solution so that the maximum absorbance is 0.7 to 1.2 AU. Molar concentrations of the NANOGOLD® and antibody are calculated from the UV/visible spectrum of the solution, and the ratio of these values gives the average number of NANOGOLD® particles per antibody. The molar extinction coefficient of NANOGOLD® at 420 nm is 155,000 M-1cm-1. Molar concentrations are calculated as follows, using a value of 230,000 M-1cm-1 for the extinction coefficient of IgG at 280 nm, where vgold, 280nm/420nm is the ratio of absorbance at 280 nm / absorbance at 420 nm for NANOGOLD®:
[NANOGOLD®] = [A420nm]/155,000
[IgG] = [A280nm vgold, 280nm/420nm x A420nm]/230,000
NANOGOLD®/antibody ratio = [NANOGOLD®]/[IgG]
To calculate the amount of antibody, A280nm is corrected for NANOGOLD® absorbtion: the absorbtion of NANOGOLD® at 280 nm is calculated by multiplying A420nm by vgold, 280nm/420nm (see product specification sheet).
Detailed instructions for this calculation are available on our web site at http://www.nanoprobes.com/LGuide4.html.
NANOGOLD® conjugates should be stored at 4°C in 0.02 M sodium phosphate buffer at pH7.4 with 150 mM sodium chloride; if the protein concentration is less than 1 mg/ml, add 0.1 % bovine serum albumin and 0.05 % sodium azide to stabilize the proteins, to prevent bacterial contamination.
Contents
If F(ab')2 fragments are available, they should be used for this purpose. If these are not available, they may be prepared from IgG molecules by pepsin7 or ficin8 digestion. Immobilized enzymes and antibody digestion kits are available for this purpose from a number of suppliers.
- Dissolve 0.15 mg of F(ab)2 (15 nmol) in 1 mL of 0.1 M sodium phosphate buffer, pH 6.0, containing 5 mM EDTA. If F(ab)2 is already in solution, exchange into the phosphate/EDTA buffer using membrane centrifugation (e.g. Centricon-30 system, Millipore) or a desalting column. Add 1 mL of the F(ab)2 solution to a vial containing 6 mg of mercaptoethylamine hydrochloride (MEA) (Final concentration is 0.05M). Incubate at 37°C for 90 minutes.
- Isolate reduced Fab' fragments by gel filtration chromatography. Use a desalting gel, such as Matrex GH25 (Millipore) which has an exclusion cutoff at molecular weight 3,000. As the eluent, use 0.02 M sodium phosphate at pH 6.5, with 150 mM sodium chloride and 1 mM EDTA. The reduced antibody will be eluted in the void volume as the first sharp peak in the trace. Combine the fractions containing reduced antibody; the total amount of antibody should be calculated from the optical density (usually for Fab', E1% at 280 nm = 15.3; concentration in mg/mL = OD280 nm X 10/E1% = OD280nm X 0.65).
- Dissolve the NANOGOLD® reagent in 0.2 mL of deionized water. If the reagent is slow to dissolve, vortex the solution. Sufficient reagent is supplied to label up to 0.15 mg of Fab'; if you are using a smaller amount, use a proportionately smaller amount of the NANOGOLD® reagent (e.g. half the reagent supplied will label 0.075 mg of Fab'). Once the NANOGOLD® reagent is reconstituted with water it should be used immediately. The maleimide group is hydrolyzed in aqueous solution.
- Add the reconstituted NANOGOLD® solution to the reduced antibody immediately. After mixing, adjust the reaction volume to give a Fab concentration of 3.5 nmol/mL (0.175 mg/mL), then incubate for 2 hours at room temperature. For an overnight reaction at 4°C, reduce the reaction volume to give a Fab concentration of 20 nmol/mL (1 mg/mL) using a membrane centrifugation device with a 10 kDa molecular weight cut-off (e.g. Centricon-10 system, Millipore).
- Separate the unbound gold particles from the antibody conjugates using gel filtration liquid chromatography. The NANOGOLD® conjugate may be effectively isolated using a medium such as GE Healthcare (Amersham Pharmacia) Superose 6 or 12 (which fractionate a wide range of molecular weights) or Superdex-75. Concentrate the reaction mixture to a suitably small volume using membrane centrifugation (e.g. Centricon-30 system, Millipore). Elute with 0.02 M sodium phosphate at pH 7.4 with 150 mM sodium chloride; the first, pale brown band is the conjugate, while the second, usually darker band is unbound NANOGOLD® particles. For even higher purity, repeat the process one time.
Estimation of NANOGOLD®/antibody ratio: Dilute a portion of the labeled antibody solution so that the maximum absorbance is 0.7 to 1.2 AU. Molar concentrations of the NANOGOLD® and antibody are calculated from the UV/visible spectrum of the solution, and the ratio of these values gives the average number of NANOGOLD® particles per antibody. The molar extinction coefficient of NANOGOLD® at 420 nm is 155,000 M-1cm-1. Molar concentrations are calculated as follows, using a value of 82,500 M-1cm-1 for the extinction coefficient of Fab' at 280 nm, where vgold, 280nm/420nm is the ratio of absorbance at 280 nm / absorbance at 420 nm for NANOGOLD®:
[NANOGOLD®] = [A420nm]/155,000
[Fab'] = [A280nm vgold, 280nm/420nm x A420nm]/82,500
NANOGOLD®/antibody ratio = [NANOGOLD®]/[Fab']
To calculate the amount of antibody, A280nm is corrected for NANOGOLD® absorbtion: the absorbtion of NANOGOLD® at 280 nm is calculated by multiplying A420nm by vgold, 280nm/420nm (see product specification sheet).
Detailed instructions for this calculation are available on our web site at http://www.nanoprobes.com/LGuide4.html.
NANOGOLD® conjugates should be stored at 4°C in 0.02 M sodium phosphate buffer at pH7.4 with 150 mM sodium chloride; if the protein concentration is less than 1 mg/ml, add 0.1 % bovine serum albumin and 0.05 % sodium azide to stabilize the proteins, to prevent bacterial contamination.
Contents
MONOMALEIMIDO NANOGOLD® may be used to label any protein with an accessible sulfhydryl group, such as a cysteine residue, in the same manner as described above for antibodies.
In some proteins the sulfhydryl functionality is in the form of a disulfide group; this must be reduced with a reducing agent, such as dithiothreitol (DTT) or mercaptoethylamine hydrochloride (MEA), before it can be labeled. If you are unsure of the structure of your protein, and have sufficient quantity available, it is recommended that the suitability of the sulfhydryl for labeling be determined first; some sulfhydryl sites may be buried within the protein structure, and therefore inaccessible to the NANOGOLD® reagent. The suitability of a particular protein for NANOGOLD® labeling may be determined using 14C iodoacetic acid before labeling is tried; alternatively, a sensitive colorimetric procedure exists for sulfhydryl determination.9
Sufficient NANOGOLD® reagent is supplied to label up to 3 nmol of sulfhydryl groups (for example, 0.3 mg of a 100,000 molecular weight compound with one sulfhydryl). Once the NANOGOLD® reagent is reconstituted with water it should be used immediately; the maleimide group is hydrolyzed in aqueous solution. The reaction should be performed in the same manner as for antibodies and antibody fragments. For purification and isolation steps alternative buffers may be substituted for those given; however, the labeling reaction itself should be performed using the buffers and conditions specified.
- If the labeling site is in the form of a disulfide group, it may be reduced to the free sulfhydryl with mercaptoethylamine hydrochloride (MEA). Dissolve the protein in 1 ml of 0.1 M sodium phosphate buffer, pH 6.0, containing 5 mM EDTA. If the protein is already in solution, perform a buffer exchange with the phosphate/EDTA buffer using membrane centrifugation (e.g. Centricon-10 system, Millipore) or a desalting column. Add 1 ml of the protein solution to the vial that contains 6 mg of mercaptoethylamine hydrochloride (MEA) (Final concentration is 0.05M). Incubate at 37°C for 90 minutes.
- Isolate reduced protein by gel filtration chromatography. Use a gel, such as Matrex GH25 (Millipore), which has an exclusion cut-off at molecular weight 3,000. Dialysis does NOT provide acceptable purification in this application. As the eluent, use 0.02 M sodium phosphate at pH 6.5, with 150 mM sodium chloride and 1 mM EDTA. The reduced protein will be eluted in the void volume as the first sharp peak in the trace. Combine the fractions containing reduced proteins; the total amount of proteins should be calculated from the optical density.
- Dissolve the NANOGOLD® reagent in 1 mL of deionized water. If the reagent is slow to dissolve, vortex the solution. The NANOGOLD® reagent can be added to the protein in 1.5 to 4-fold mole excess. Once the NANOGOLD® reagent is reconstituted with water it should be used immediately. The maleimide group is hydrolyzed in aqueous solution.
If solubility is known to be an issue with the molecule you are labeling, solubility may be improved by predissolving the NANOGOLD® reagent in up to 5 or 10% final volume (0.01 to 0.02 mL) of ispropanol, then making up to 0.2 mL with water.
- Add the reconstituted NANOGOLD® solution to the reduced protein. Adjust the reaction volume to give a protein concentration of 3 to 6 nmol/mL (for a 100,000 MW protein, this is 0.3 to 0.6 mg/mL). If a membrane centrifugation unit is used to reduce the volume of the reaction mixture, the molecular weight cut-off limit of 10,000 (e.g. Centricon-10 system, Millipore) should be used to avoid losing Nanogold reagent. Incubate the reaction mixture for 2 hours at room temperature, or overnight at 4°C.
- Separate the unbound gold particles from the protein conjugates using gel filtration chromatography. The NANOGOLD® conjugate may be effectively isolated using a medium such as GE Healthcare (Amersham Pharmacia) Superose 6 or 12 (which fractionate a wide range of molecular weights) or Superdex-75. Concentrate the reaction mixture to a suitably small volume using membrane centrifugation (e.g. Centricon-30 system, Millipore). Elute with 0.02 M sodium phosphate at pH 7.4 with 150 mM sodium chloride; the first, pale brown band is the conjugate, while the second, usually darker band is unbound NANOGOLD® particles. For even higher purity, repeat the process one time.
Estimation of NANOGOLD®/protein ratio: Dilute a portion of the labeled protein solution so that the maximum absorbance is 0.7 to 1.2 AU. Molar concentrations of the NANOGOLD® and protein are calculated from the UV/visible spectrum of the solution, and the ratio of these values gives the average number of NANOGOLD® particles per antibody. The molar extinction coefficient of NANOGOLD® at 420 nm is 155,000 M-1cm-1. Molar concentrations are calculated as follows, using a value of 120,000 M-1cm-1 for the extinction coefficient of protein at 280 nm, where vgold, 280nm/420nm is the ratio of absorbance at 280 nm / absorbance at 420 nm for NANOGOLD®. The extinction coefficient will vary for different proteins; it may be calculated from the molecular weight and optical density.
[NANOGOLD®] = [A420nm]/155,000
[protein] = [A280nm vgold, 280nm/420nm x A420nm]/120,000
NANOGOLD®/protein ratio = [NANOGOLD®]/[protein]
To calculate the amount of protein, A280nm is corrected for NANOGOLD® absorbtion: the absorbtion of NANOGOLD® at 280 nm is calculated by multiplying A420nm by vgold, 280nm/420nm (see product specification sheet).
Detailed instructions for this calculation are available on our web site at http://www.nanoprobes.com/LGuide4.html.
NANOGOLD® conjugates should be stored at 4°C in 0.02 M sodium phosphate buffer at pH7.4 with 150 mM sodium chloride, or other buffer solutions usually used with the protein under study. If the conjugates are intended for long storage, add 0.1 % bovine serum albumin and 0.05 % sodium azide to prevent bacterial contamination and to prevent the protein from adhering to the surfaces of the storage vessel.
Contents
MONOMALEIMIDO NANOGOLD® may be used in the same manner to label smaller proteins and peptides. However, for a protein or peptide which is smaller than NANOGOLD®, the ratios of NANOGOLD® to protein are reversed (i.e. an excess of protein should be used): an excess of the most easily separated component (usually the smallest) should be used, because this is more easily separated from the product that unbound NANOGOLD®. The molecular weight of NANOGOLD® is close to 15,000, but because of the contribution from high Z gold atoms, it is smaller than a protein of equal mass: including its coordinated ligands, NANOGOLD® is about 2.6 nm in diameter, equivalent to a globular protein of molecular weight 8,000 to 10,000.
If the sulfhydryl functionality is in the form of a disulfide group, it must be reduced to a thiol for labeling. If a thiol-based reducing agent such as dithiothreitol (DTT), mercaptoethylamine hydrochloride (MEA) or mercaptoethanol is used it should be removed by gel filtration chromatography before mixing with the NANOGOLD® reagent. If you are unsure of the structure of your protein, and have sufficient quantity available, it is recommended that the suitability of the sulfhydryl for labeling be determined first; some sulfhydryl sites may be buried within the protein structure, and therefore inaccessible to the NANOGOLD reagent. The suitability of a particular protein for NANOGOLD® labeling may be determined using 14C iodoacetic acid before labeling is tried; alternatively, a sensitive colorimetric procedure exists for sulfhydryl determination.9
Sufficient NANOGOLD® reagent is supplied to label up to 6 nmol of sulfhydryl groups (for example, 0.03 mg of a 5,000 molecular weight compound with one sulfhydryl). A two-fold to ten-fold excess of the protein or peptide should be used (smaller excesses for peptides closer to the MW of NANOGOLD®); for the example above, this is about 0.15 0.3 mg. Once the NANOGOLD® reagent is reconstituted with water it should be used immediately; the maleimide group is hydrolyzed in aqueous solution. For purification and isolation steps alternative buffers may be substituted for those given; however, the labeling reaction itself should be performed using the buffers and conditions specified.
- If the labeling site is in the form of a disulfide group, it may be reduced to the free sulfhydryl with mercaptoethylamine hydrochloride (MEA). Dissolve the protein in 1 ml of 0.1 M sodium phosphate buffer, pH 6.0, containing 5 mM EDTA. If the protein is already in solution, exchange into the phosphate/EDTA buffer using membrane centrifugation (e.g. Centricon-30 system, Millipore) or a desalting column. Add 1 mL of the protein solution to the vial that contains 6 mg of mercaptoethylamine hydrochloride (MEA) (Final concentration is 0.05M). Incubate at 37°C for 90 minutes.
- Isolate reduced protein or peptide by gel filtration chromatography. Use a desalting gel (such as Matrex GH25, Millipore) which has an exclusion cut-off at molecular weight lower than that of the protein or peptide. Dialysis does NOT provide acceptable purification in this application. As the eluent, use 0.02 M sodium phosphate at pH 6.5, with 150 mM sodium chloride and 1 mM EDTA. The reduced protein will be eluted in the void volume as the first sharp peak in the trace. Combine the fractions containing reduced protein; the total amount of protein should be calculated from the optical density.
- Dissolve the NANOGOLD® reagent in 0.2 mL of deionized water. If the reagent is slow to dissolve, vortex the solution. Smaller volumes are not recommended since the reagent contains buffers. 30 nmol of reagent is supplied: this will react with 0.03 mg of a 5,000 MW peptide containing one sulfhydryl. If you are using a smaller amount, use a proportionately smaller amount of the NANOGOLD® reagent (e.g. half the reagent supplied will label up to 0.015 mg of the above peptide). Once the NANOGOLD® reagent is reconstituted with water it should be used immediately. The maleimide group is hydrolyzed in aqueous solution.
If water-solubility is known to be an issue with the molecule you are labeling, solubility may be improved by predissolving the NANOGOLD® reagent in up to 5 or 10% final volume (0.01 to 0.02 mL) of ispropanol, then making up to 0.2 mL with water.
In addition to aqueous solubility, NANOGOLD® reagents are also highly soluble in DMSO, and this may be used as a solvent for labeling molecules such as certain peptides that are insoluble in water but soluble in organic solvents. Dissolve the NANOGOLD® reagent in 1 ml of anhydrous DMSO; some undissolved buffer salts may remain, but these will not affect the labeling reaction. Mix with the molecule to be labeled, dissolved in an anhydrous organic solvent. Overnight stirring is normally necessary for reaction to be complete.
- Add the activated NANOGOLD® solution to the reduced protein or peptide. Adjust the reaction volume to give a peptide concentration of 3 to 6 nmol/mL (for a 5,000 MW peptide, this is 0.015 to 0.03 mg/mL). If a membrane centrifugation unit is used to reduce the volume of the reaction mixture, the molecular weight cut-off limit less than 6,000 (e.g. Centricon-3 system, Millipore) should be used to avoid losing protein or peptide or Nanogold reagent. Incubate the reaction mixture for 2 hours at room temperature, or overnight at 4°C.
- Separate the unbound gold particles from the protein conjugates using gel filtration chromatography. The NANOGOLD® conjugate may be effectively isolated using a medium such as GE Healthcare (Amersham Pharmacia) Superdex-75 or Superdex-Peptide, or Bio-Rad Bio-Gel P2, P4, P6 or P10 (which fractionate a wide range of molecular weights), or for very small peptides, GH25 (Millipore; excludes molecules larger than MW 3,000). Concentrate the reaction mixture to a suitably small volume using membrane centrifugation (e.g. Centricon-3 or 5 system, Millipore). Elute with 0.02 M sodium phosphate at pH 7.4 with 150 mM sodium chloride; the first, colored peak is the conjugate, while the second, less colored band is unbound protein or other smaller molecules. For even higher purity, repeat the process one time.
Estimation of NANOGOLD®/peptide ratio: Dilute a portion of the labeled peptide solution so that the maximum absorbance is 0.7 to 1.2 AU. Molar concentrations of the NANOGOLD® and peptide are calculated from the UV/visible spectrum of the solution, and the ratio of these values gives the average number of NANOGOLD® particles per antibody. The molar extinction coefficient of NANOGOLD® at 420 nm is 155,000 M-1cm-1. Molar concentrations are calculated as follows, using a value of 12,000 M-1cm-1 for the extinction coefficient of peptide at 280 nm, where vgold, 280nm/420nm is the ratio of absorbance at 280 nm / absorbance at 420 nm for NANOGOLD®. The extinction coefficient will vary for different proteins; it may be calculated from the molecular weight and optical density.
[NANOGOLD®] = [A420nm]/155,000
[peptide] = [A280nm vgold, 280nm/420nm x A420nm]/12,000
NANOGOLD®/peptide ratio = [NANOGOLD®]/[peptide]
To calculate the amount of peptide, A280nm is corrected for NANOGOLD® absorbtion: the absorbtion of NANOGOLD® at 280 nm is calculated by multiplying A420nm by vgold, 280nm/420nm (see product specification sheet).
Detailed instructions for this calculation are available on our web site at http://www.nanoprobes.com/LGuide4.html.
NANOGOLD® conjugates should be stored at 4°C in 0.02 M sodium phosphate buffer at pH7.4 with 150 mM sodium chloride, or other buffer solutions usually used with the peptide under study. If the conjugates are intended for long storage, add 0.1 % bovine serum albumin and 0.05 % sodium azide to prevent bacterial contamination and to prevent the protein from adhering to the surfaces of the storage vessel.
Note: Proteins with molecular weights between 6,000 and 15,000 should be labeled using the same procedure; however, a more nearly stoichiometrically equal amount of NANOGOLD® should be used. If possible, use a slight excess (2-fold) of the component most easily separated (depending on the nature of the protein and the methods usually used for separation, such as ion exchange chromatography of hydrophobic interaction chromatography, this might be the protein or the NANOGOLD®).
Contents
Basically, normal methodologies may be used successfully with NANOGOLD® immunoreagents. The concentration of antibody and gold is similar to other commercial preparations of colloidal gold antibodies. Therefore similar dilutions and blocking agents are appropriate.
The major difference will be in the results:
- NANOGOLD® is an extremely uniform 1.4 nm diameter gold particle (ca.10%).
- NANOGOLD® conjugates contain absolutely no aggregates. This is in sharp contrast to other colloidal gold conjugates that usually are prepared by centrifugation to remove the largest aggregates and frequently contain smaller aggregates.
- Close to 1 NANOGOLD® particle to 1 Fab (or IgG) make this product distinct from the 0.2 - 10 variable stoichiometry of other colloidal gold antibody preparations.
- NANOGOLD® particles do not have affinity to proteins as do other other colloidal golds. This reduces background and false labeling.
- NANOGOLD® develops better with silver than do most other colloidal golds giving it higher sensitivity. Silver enhancement can be used to make the immunolabeling useful for light microscopy and immunoblotting with improved results.
Contents
Because the 1.4 nm NANOGOLD® particles are so small, over staining with OsO4, uranyl acetate or lead citrate may tend to obscure direct visualization of individual NANOGOLD® particles. Three recommendations for improved visibility of NANOGOLD® are:
- Use of reduced amounts or concentrations of usual stains.
- Use of lower atomic number stains such as NANOVAN, a Vanadium based stain.10
- Enhancement of NANOGOLD® with silver developers, such as LI SILVER or HQ SILVER.
Contents
If aldehyde-containing reagents have been used for fixation, these must be quenched before labeling. This may be achieved by incubating the specimens for 5 minutes in 50 mM glycine solution in PBS (pH 7.4). Ammonium chloride (50 mM) or sodium borohydride (0.5 - 1 mg/ml) in PBS may be used instead of glycine (see Ref. 4 for further details).
Cells in Suspension
- Optional fixing of cells: e.g., with glutaraldehyde (0.05 - 1% for 15 minutes) in PBS. Do not use Tris buffer since this contains an amine. After fixation, centrifuge cells (e.g. 1 ml at 10,000,000 cells/ml) at 300 X g, 5 minutes; discard supernatant; resuspend in 1 ml buffer. Repeat this washing (centrifugation and resuspension) 2 times.
- Incubate cells with 0.02 M glycine in PBS (5 mins). Centrifuge, then resuspend cells in PBS-BSA buffer (specified below) for 5 minutes.
- Wash cells using PBS-BSA as described in step 1 (2 X 5 mins). Resuspend in 1 ml Buffer 1.
- Place 50 - 200 microliters of cells into Eppendorf tube. Dilute NANOGOLD® conjugate ~ 50 times in PBS-BSA buffer and add 30 microliters to cells; incubate for 30 minutes with occasional shaking (do not create bubbles which will denature proteins).
- Wash cells in PBS-BSA as described in step 1 (2 X 5 mins).
- Fix cells and antibodies using a final concentration of 1% glutaraldehyde in PBS for 15 minutes. Then remove fixative by washing with buffer 1 (3 X 5 mins).
PBS-BSA Buffer:
20 mM phosphate
150 mM NaCl
pH 7.40
0.5% BSA
0.1% gelatin (high purity)
Optional, may reduce background:
0.5 M NaCl
0.05% Tween 20
|
PBS Buffer:
20 mM phosphate
150 mM NaCl
pH 7.40
|
Negative staining may be used for electron microscopy of small structures or single molecules which are not embedded. Negative stain must be applied after the silver enhancement. NANOVAN negative stain is specially formulated for use with NANOGOLD® reagents; it is based on vanadium, which gives a lighter stain than uranium, lead or tungsten-based negative stains and allows easier visualization of NANOGOLD® particles with little or no silver enhancement.10
CAUTION: NANOGOLD® particles degrade upon exposure to concentrated thiols such as beta-mercaptoethanol or dithiothreitol. If such reagents must be used, concentrations should be kept below 1 mM and exposure restricted to 10 minutes or less.
Thin Sections
Labeling with NANOGOLD® may be performed before or after embedding.11,12 Labeling before embedding and sectioning (the pre-embedding method)11,12 is used for the study of surface antigens, particularly small organisms such as viruses budding from host cells. It gives good preservation of cellular structure, and subsequent staining usually produces high contrast for study of the cellular details. Labeling after embedding and sectioning (the post-embedding method)11,12 allows the antibody access to the interior of the cells, and is used to label both exterior and interior features. The procedures for both methods are described below.
Thin sections mounted on grids are floated on drops of solutions on parafilm or in well plates. Hydrophobic resins usually require pre-etching.
CAUTION: Nanogold® particles degrade upon exposure to concentrated thiols such as beta-mercaptoethanol or dithiothreitol. If such reagents must be used, concentrations should be kept below 1 mM and exposure restricted to 10 minutes or less. NANOGOLD® particles are not stable above 37°C. If immunolabeling is performed before embedding, an embedding medium should be used that hardens at low temperature (<30°C, e.g. Lowicryl). See temperature caution above.
PROCEDURE FOR PRE-EMBEDDING METHOD:11
- Float on a drop of water for 5 - 10 minutes.
- Incubate cells with 1 % bovine serum albumin in PBS buffer at pH 7.4 for 5 minutes; this blocks any non-specific protein binding sites and minimizes non-specific antibody binding.
- Rinse with PBS-BSA (1 min).
- Incubate with NANOGOLD® conjugate diluted 1/40 - 1/200 in PBS-BSA with 1 % normal serum from the same species as the NANOGOLD® labeled antibody, for 10 minutes to 1 hour at room temperature.
- Rinse with PBS-BSA (3 X 1 min), then PBS (3 X 1 min).
- Postfix with 1 % glutaraldehyde in PBS (10 mins).
- Rinse in deionized water (2 X 5 min).
- Dehydrate and embed according to usual procedure. Use of a low-temperature resin (eg. Lowicryl) is recommended.
- Stain (uranyl acetate, lead citrate or other positive staining reagent) as usual before examination.
Silver enhancement may be performed before or after embedding (see below); it should be completed before postfixing or staining with osmium tetroxide, uranyl acetate or similar reagents is carried out.
PROCEDURE FOR POST-EMBEDDING METHOD:11
- Prepare sections on plastic or carbon-coated nickel grid. Float on a drop of water for 5 - 10 minutes.
- Incubate with 1 % solution of bovine serum albumin in PBS buffer at pH 7.4 for 5 minutes to block non-specific protein binding sites.
- Rinse with PBS-BSA (1 min).
- Incubate with NANOGOLD® conjugate diluted 1/40 - 1/200 in PBS-BSA with 1 % normal serum from the same species as the NANOGOLD® labeled antibody, for 10 minutes to 1 hour at room temperature.
- Rinse with PBS (3 X 1 min).
- Postfix with 1 % glutaraldehyde in PBS at room temperature (3 mins).
- Rinse in deionized water for (2 X 5 min).
- If desired, contrast sections with uranyl acetate and/or lead citrate before examination.
Silver enhancement may also be used to render the NANOGOLD® particles more easily visible (see below), especially if stains such as uranyl acetate or lead citrate are applied. If used, it should be completed before these satins are applied.
PBS-BSA Buffer:
20 mM phosphate
150 mM NaCl
pH 7.40
0.5% BSA
0.1% gelatin (high purity)
Optional, may reduce background:
0.5 M NaCl
0.05% Tween 20
|
PBS Buffer:
20 mM phosphate
150 mM NaCl
pH 7.40
|
Contents
For most work, silver enhancement is recommended to give a good signal in the electron microscope (see below). For particular applications, visualization of the NANOGOLD® directly may be desirable. Generally this requires very thin samples and precludes the use of other stains.
NANOGOLD® provides a much improved resolution and smaller probe size over other colloidal gold antibody products. However, because NANOGOLD® is only 1.4 nm in diameter, it will not only be smaller, but will appear less intense than, for example, a 5 nm gold particle. With careful work, however, NANOGOLD® may be seen directly through the binoculars of a standard EM even in 80 nm thin sections. However, achieving the high resolution necessary for this work may require new demands on your equipment and technique. Several suggestions follow:
- Before you start a project with NANOGOLD® it is helpful to see it so you know what to look for. Dilute the NANOGOLD® stock 1:5 and apply 4 ml to a grid for 1 minute. Wick the drop and wash with deionized water 4 times.
- View NANOGOLD® at 100,000 X magnification with 10 X binoculars for a final magnification of 1,000,000 X. Turn the emission up full and adjust the condenser for maximum illumination.
- The alignment of the microscope should be in order to give 0.3 nm resolution. Although the scope should be well aligned, you may be able to skip this step if you do step 4
- Objective stigmators must be optimally set at 100,000 X. Even if the rest of the microscope optics are not perfectly aligned, adjustment of the objective stigmators may compensate and give the required resolution. You may want to follow your local protocol for this alignment but since it is important, a brief protocol is given here:
- At 100,000 X (1 X 106 with binoculars), over focus, under focus, then set the objective lens to in focus. This is where there is the least amount of detail seen.
- Adjust each objective stigmator to give the least amount of detail in the image.
- Repeat steps a and b until the in focus image contains virtually no contrast, no wormy details, and gives a flat featureless image.
- Now underfocus slightly, move to a fresh area, and you should see small black dots of 1.4 nm size. This is the NANOGOLD®. For the 1:5 dilution suggested, there should be about 5 to 10 gold spots on the small viewing screen used with the binoculars. Contrast and visibility of the gold clusters is best at 0.2 - 0.5 m defocus, and is much worse at typical defocus values of 1.5 - 2.0 m commonly used for protein molecular imaging.
- In order to operate at high magnification with high beam current, thin carbon film over fenestrated holey film is recommended. Alternatively, thin carbon or 0.2% Formvar over a 1000 mesh grid is acceptable. Many plastic supports are unstable under these conditions of high magnification/high beam current and carbon is therefore preferred. Contrast is best using thinner films and thinner sections.
- Once you have seen NANOGOLD® you may now be able to reduce the beam current and obtain better images on film. For direct viewing with the binoculars reduction in magnification from 1,000,000 X to 50,000 X makes the NANOGOLD® much more difficult to observe and not all of the golds are discernable. At 30,000 X (300,000 X with 10 X binoculars) NANOGOLD® particles are not visible. It is recommended to view at 1,000,000 X, with maximum beam current, align the objective stigmators, and then move to a fresh area, reduce the beam, and record on film.
- If the demands of high resolution are too taxing or your sample has an interfering stain, a very good result may be obtained using silver enhancement to give particles easily seen at lower magnification.
Contents
NANOGOLD® will nucleate silver deposition resulting in a dense particle 2-80 nm in size or larger depending on development time. If specimens are to be embedded, silver enhancement is usually performed after embedding, although it may be done first. It must be completed before any staining reagents such as osmium tetroxide, lead citrate or uranyl acetate are applied, since these will nucleate silver deposition in the same manner as gold and produce non-specific staining. With NANOGOLD® reagents, low-temperature resins (eg Lowicryl) should be used and the specimens kept at or below room temperature until after silver development has been completed. Silver development is recommended for applications of NANOGOLD® in which these stains are to be used, otherwise the NANOGOLD® particles may be difficult to visualize against the stain.
Our LI SILVER silver enhancement system is convenient and not light sensitive, and suitable for all applications. Improved results in the EM may be obtained using HQ SILVER, which is formulated to give slower, more controllable particle growth and uniform particle size distribution.13
Specimens must be thoroughly rinsed with deionized water before silver enhancement reagents are applied. This is because the buffers used for antibody incubations and washes contain chloride ions and other anions which form insoluble precipitates with silver. These are often light-sensitive and will give non-specific staining. To prepare the developer, mix equal amounts of the enhancer and initiator immediately before use. NANOGOLD® will nucleate silver deposition resulting in a dense particle 2-20 nm in size or larger depending on development time. Use nickel grids (not copper).
The procedure for immunolabeling should be followed up to step 6 as described above. Silver enhancement is then performed as follows:
- Rinse with deionized water (2 X 5 mins).
- OPTIONAL (may reduce background): Rinse with 0.02 M sodium citrate buffer, pH 7.0 (3 X 5 mins).
- Float grid with specimen on freshly mixed developer for 1-8 minutes, or as directed in the instructions for the silver reagent. More or less time can be used to control particle size. A series of different development times should be tried, to find the optimum time for your experiment. With HQ Silver, a development time of 6 mins gives 15-40 nm round particles.
- Rinse with deionized water (3 X 1 min).
- Mount and stain as usual.
Fixing with osmium tetroxide may cause some loss of silver; if this is found to be a problem, slightly longer development times may be appropriate.
NOTE: Treatment with osmium tetroxide followed by uranyl acetate staining can lead to much more drastic loss of the silver enhanced NANOGOLD® particles. This may be prevented by gold toning:14
- After silver enhancement, wash thoroughly with dionized water.
- 0.05 % gold chloride: 10 minutes at 4°C.
- Wash with deionized water.
- 0.5 % oxalic acid: 2 mins at room temperature.
- 1 % sodium thiosulfate (freshly made) for 1 hour.
- Wash thoroughly with deionized water and embed according to usual procedure.
Contents
Features labeled with NANOGOLD® will be stained black in the light microscope upon silver enhancement. Different development times should be tried to determine which is best for your experiment. The procedure for immunolabeling is similar to that for EM; a suitable procedure is given below.
Samples must be rinsed with deionized water before silver enhancement. This is because the reagent contains silver ions in solution, which react to form a precipitate with chloride, phosphate and other anions which are components of buffer solutions. The procedure for immunolabeling with NANOGOLD® and silver enhancement is given overleaf.
- Spin cells onto slides using Cytospin, or use parafin section.
- Incubate with 1 % solution of bovine serum albumin in PBS (PBS-BSA) for 10 minutes to block non-specific protein binding sites.
- Rinse with PBS-BSA (3 X 2 min).
- Incubate with NANOGOLD® reagent diluted 1/40 - 1/200 in PBS-BSA with 1 % normal serum from the same species as the NANOGOLD® reagent, for 1 hour at room temperature.
- Rinse with PBS (3 X 5 min).
- Postfix with 1 % glutaraldehyde in PBS at room temperature (3 mins).
- Rinse with deionized water (3 X 1 min).
- OPTIONAL (may reduce background): Rinse with 0.02 M sodium citrate buffer, pH 7.0 (3 X 5 mins).
- Develop specimen with freshly mixed developer for 5-20 minutes, or as directed in the instructions for the silver reagent. More or less time can be used to control intensity of signal. A series of different development times may be used, to find the optimum enhancement for your experiment; generally a shorter antibody incubation time will require a longer silver development time.
- Rinse with deionized water (2 X 5 mins).
- The specimen may now be stained if desired before examination, with usual reagents.
PBS-BSA Buffer:
20 mM phosphate
150 mM NaCl
pH 7.40
0.5% BSA
0.1% gelatin (high purity)
Optional, may reduce background:
0.5 M NaCl
0.05% Tween 20
|
PBS Buffer:
20 mM phosphate
150 mM NaCl
pH 7.40 |
Contents
The basic procedure for gold immunoblotting has been described by Moeremans et al,15 which may be followed. For best results, the membrane should be hydrated before use by simmering in gently boiling water for 15 minutes. Best results are obtained when the antigen is applied using a 1 microliter capillary tube. The procedure for immunoblots is as follows, if the NANOGOLD® conjugate is the primary antibody:
- Spot 1 microliter dilutions of the antigen in buffer 4 onto hydrated nitrocellulose membrane. Use an antigen concentration range from 100 to 0.01 pg / microliter.
- Block with buffer 1 for 30 minutes at 37°C.
- Rinse with buffer 1 (3 X 10 mins).
- Incubate with a 1/100 to 1/200 dilution of the NANOGOLD® reagent in buffer 2 for 2 hours at room temperature.
- Rinse with buffer 3 (3 X 5 mins), then buffer 4 (2 X 5 mins).
- OPTIONAL (may improve sensitivity): Postfix with glutaraldehyde, 1 % in buffer 4 (10 mins).
- Rinse with deionized water (2 X 5 mins).
- OPTIONAL (may reduce background): Rinse with 0.05 M EDTA at pH 4.5 (5 mins).
- Develop with freshly mixed silver developer for 20-25 minutes or as directed in the instructions for the silver reagent, twice. Rinse thoroughly with deionized water between developments to remove all the reagent.
- Rinse several times with deionized water.
CAUTION: NANOGOLD® particles degrade upon exposure to concentrated thiols such as beta-mercaptoethanol or dithiothreitol. If such reagents must be used, concentrations should be kept below 1 mM and exposure restricted to 10 minutes or less.
Buffer 1:
20 mM phosphate
150 mM NaCl
pH 7.40
4% BSA (bovine serum albumin)
2 mM sodium azide (NaN3)
|
Buffer 2:
20 mM phosphate
150 mM NaCl
pH 7.4
0.8% BSA
1% normal serum; use serum of the host animal for the NANOGOLD® antibody
0.1% gelatin (Type B, approx. 60 bloom)
Optional, may reduce background:
0.5 M NaCl
0.05% Tween 20
|
Buffer 3:
20 mM phosphate
150 mM NaCl
pH 7.40
0.8% BSA (bovine serum albumin)
2 mM sodium azide
|
Buffer 4 (PBS):
20 mM phosphate
150 mM NaCl
pH 7.4
|
Other procedures may be used; for example the NANOGOLD® reagent may be used as a tertiary labeled antibody, or a custom NANOGOLD® conjugate may be the primary antibody. If additional antibody incubation steps are used, rinse with buffer 3 (3 X 10 mins) after incubation.
Contents
- Hainfeld, J. F., and Powell R. D.: Nanogold Technology: New Frontiers in Gold Labeling. Cell Vision, 4, 308-324 (1997); Furuya, F. R., and Hainfeld, J. F.: A 1.4-nm gold cluster covalently attached to antibodies improves immunolabeling. J. Histochem. Cytochem., 40, 177 (1992); Furuya, F. R., Hainfeld, J. F., and Powell, R. D.: A new 1.4 nm Gold-Fab' Probe. Proc. 49th Ann. Mtg., Electron.Micros. Soc. Amer.; Bailey, G. W. (Ed.), San Francisco Press, San Francisco, CA; 1991, p. 284.
- Boisset, N., Penczek, P., Pochon, F., Frank, J., and Lamy, J.: Three-dimensional reconstruction of human alpha 2-macroglobulin and refinement of the localization of thiol ester bonds with monomaleimido nanogold. Ann. NY Acad. Sci., 737, 229-244 (1994).
- Gregori, L., Hainfeld, J. F., Simon, M. N., and Goldgaber, D.: Binding of amyloid beta protein to the 20 S proteasome. J. Biol. Chem., 272, 58-62 (1997).
- Alivisatos, A. P., Johnsson, K. P., Peng, X., Wilson, T. E., Loweth, C. J., Bruchez, M. P., Jr., and Schultz, P. G.: Organization of 'nanocrystal molecules' using DNA. Nature, 382, 609-611 (1996).
- Hainfeld, J. F., and Furuya, F. R.; in Immunogold-Silver Staining: Principles, Methods and Applications (M. A. Hayat, Ed.), CRC Press, Boca raton, FL., 1995: pp. 71-96.
- Krencs, T., and Krencs, L.; in Immunogold-Silver Staining: Principles, Methods and Applications (M. A. Hayat, Ed.), CRC Press, Boca raton, FL., 1995: pp. 57-69.
- Mariani, M.; Camagna, M.; Tarditi, L., and Seccamani, E.: A new enzymatic method to obtain high-yield F(ab)2 suitable for clinical use from mouse IgGl. Molecular Immunology, 28, 69-77 (1991).
- Parham, P.: On the fragmentation of monoclonal IgG1, IgG2a, and IgG2b from BALB/c mice. J. Immunology, 131, 2895-2902 (1983).
- Grassetti, D. R., and Murray, J. F. Jr.; Arch. Biochem. Biophys., 119, 41 (1967).
- Tracz, E., Dickson, D. W., Hainfeld, J. F., and Ksiezak-Reding, H.: Paired helical filaments in corticobasal degeneration: the fine fibrillary structure with NanoVan. Brain Res., 773, 33-44 (1997); Gregori, L., Hainfeld, J. F., Simon, M. N., and Goldgaber, D. Binding of amyloid beta protein to the 20S proteasome. J. Biol. Chem., 272, 58-62 (1997); Hainfeld, J. F.; Safer, D.; Wall, J. S.; Simon, M. N.; Lin, B. J., and Powell, R. D.; Methylamine Vanadate (NanoVan) Negative Stain. Proc. 52nd Ann. Mtg., Micros. Soc. Amer.; G. W. Bailey and Garratt-Reed, A. J., (Eds.); San Francisco Press, San Francisco, CA, 1994, p. 132.
- J. E. Beesley, in Colloidal Gold: Principles, Methods and Applications, M. A. Hayat, ed., Academic Press, New York, 1989; Vol. 1, pp. 421-425.
- Lujan, R.; Nusser, Z.; Roberts, J. D. B.; Shigemoto, R.; Ohishi, H., and Somogyi, P.: Differential plasma membrane distribution of metabotropic glutamate receptors mGluR1 alpha, mGluR2 and mGluR5, relative to neurotransmitter release sites. J. Chem. Neuroanat., 13, 219-241 (1997).
- Humbel, B. M.; Sibon, O. C. M.; Stierhof, Y.-D., and Schwarz, H.: Ultra-small gold particles and silver enhancement as a detection system in immunolabeling and In Situ hybridization experiments; J. Histochem. Cytochem., 43, 735-737 (1995).
- Arai, R.; Geffard, M., and Calas, A.: Intensification of labelings of the immunogold silver staining method by gold toning. Brain Res. Bull., 28, 343-345 (1992).
- Moeremans, M.; Daneels, G.; Van Dijck, A.; Langanger, G., and De Mey, J.: Sensitive visualization of antigen-antibody reactions in dot and blot immune overlay assays with immunogold and immunogold/silver staining., J. Immunol. Meth. 74, 353-360 (1984).
Questions? Technical Support is available. Drop us a line-- we're here to help.
 |
Nanoprobes, Incorporated
95 Horseblock Road, Unit 1, Yaphank, NY 11980-9710
Tel: (877) 447-6266 (Toll-Free in US) or (631) 205-9490 | Fax: (631) 205-9493
tech@nanoprobes.com | www.nanoprobes.com |
For a complete list of references citing this product, please visit our References page.
Contents
|